Mechanisms of DNA Repair and Carcinogenesis
This team belongs to the UMR 9019 Genome Integrity and Cancers.
The research activities of the Team aim to decipher the mechanisms of repair of complex DNA damage in order to get insight into development of cancer and particularly renal one and the mechanisms of chemoresistance of tumor cells by using the complementarity of various expertise of the members of Team. Insights from mechanistic studies of the alternative DNA repair pathways and the post-replicative modifications of DNA would help to develop novel diagnostic and therapeutic targets to design repair inhibitors to combat acquired resistance in cancers, particularly in renal carcinoma which can be used as a model.
Research topics
The team “Mechanisms of DNA Repair and Carcinogenesis” studies various aspects of the enzymology of repair of endogenous and exogenous DNA damage in the cells and its implication in the carcinogenesis and resistance of cancer cells (intrinsic or acquired) to therapies, with particular attention to the renal cancer. The clinical features of certain inherited human DNA repair deficient disorders point to complex nature of endogenous oxidative DNA damage which include bulky adducts, inter-strand DNA crosslinks (ICLs) and clustered lesions. On the other hand, severe biological effects of ionizing radiation and DNA cross-linking agents are correlated with formation of complex lesions in cellular DNA. Genetic and biochemical data indicate that the elimination of complex DNA lesions requires several distinct DNA repair pathways such as base excision repair (BER), nucleotide incision repair (NIR), nucleotide excision repair, mismatch repair, translesion DNA synthesis, homologous recombination and non-homologous end-joining.
Patients with inoperable and early metastasizing tumors such as breast and lung cancers are frequently treated by the combination of poly-chemotherapy and radio-therapy. While cancer cells are primarily chemo- and radio-sensitive, they can develop over the subsequent 3 to 12 months acquired chemo- and radio-resistance. Therefore, acquired resistance of tumor cells to the treatments remains a fundamental barrier to attaining maximal efficacy with chemo- and radiotherapy for advanced cancers. Particularly, clear cell Renal Cell Carcinoma (ccRCC) is characterized by intra-tumoral hypoxia and chemoresistance. Conventional chemotherapeutic agents are ineffective to treat ccRCC, because of unusual resistance of the tumor cells to DNA damage, likely due to activated DNA repair and signaling systems. Thus, renal cancer offers us a model to study the role of DNA repair in both the genesis and resistance to anticancer therapies of ccRCC. It should be noted that the hypoxia-inducible transcription factors HIF1α and HIF2α play a crucial role in ccRCC initiation and progression. Up to one hundred genes are overexpressed when HIFs are stabilized in ccRCC, leading to tumour growth, neovascularization, invasion and metastasis. HIF is remarkably knitted together with DNA-damage response factors.
At present, the mechanisms leading to acquired resistance in cancer are not yet fully understood. Among putative mechanisms, the activation of DNA repair pathways is an attractive hypothesis. The clinical development of drugs targeting DNA repair as anticancer compounds has been designed around the concept of synthetic lethality, in which effectiveness is dependent on the genetic background of the cells with respect to competency of the repair pathways. Inhibitors of the poly(ADP-ribose) polymerases (PARP) represent the most well-known paradigm of synthetic lethality. For example, PARP1 inhibition has been shown to be most effective in tumor cells with defective homologous recombination, due to mutations in the BRCA-1 and BRCA-2 genes. Thus, understanding the mechanisms of DNA repair pathways will bring new insights into acquired resistance of tumor cells and help to develop new strategies to combat cancers.
The general objective of the Team is to explore the molecular mechanisms of the DNA damage and repair linked to carcinogenesis and chemo/radio-resistance. There are two major questions that we focus on:
- the molecular mechanisms and the role of new alternative DNA repair pathways, previously identified by our Team, in the therapeutic resistance of cancer cells,
- the role of repair of complex DNA lesions in familial cancer syndromes.
To address these questions, our Team conducts research on the three axes described below:
Axis 1: Identification and characterization of the alternative DNA repair pathways for complex DNA lesions which are implicated in malignant transformation and resistance to anti-cancer treatments.
The processing of complex DNA damage typically involves several overlapping DNA repair pathways, which should be coordinated in a spatial and orderly manner. Here, we propose that complex DNA lesions could be repaired via different pathways depending on the cell type and nature of the lesion. Previously, we established several new alternative DNA repair mechanisms of complex DNA lesions:
- Excision of DNA-DNA and DNA-protein crosslinks by human NEIL1 & NEIL3 DNA glycosylases (see Figure 1);
- Removal of bulky 8,5'-cyclo-2’-deoxypurines residues by human APE1 protein;
- Aberrant error-prone repair of oxidative DNA base damage by human thymine-DNA glycosylases (TDG & MBD4). Although these alternative repair mechanisms have the advantage of being quick and simple, using few steps and little energy, as compared to the classic regular DNA repair pathways, they can also lead to genetic instability. Indeed, chemo- and radio-resistant tumor cells are characterized by genome instability and by over-expression of APE1, NEIL3 and PARP1.
In this Axis, we address two major questions using renal cancer (ccRCC) as a model:
- Whether new non-elucidated repair activities are present in resistant cancer cells;
- What are the mechanisms of DNA repair coordination and whether the acquired resistance of cancer cells depends on the specific protein-protein interactions and post-translational modifications (PTMs).
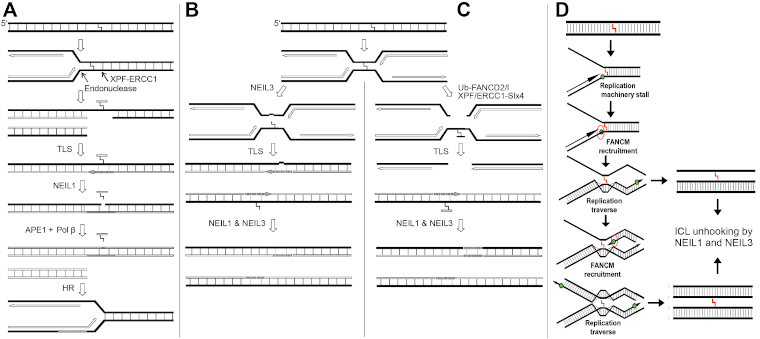
Figure 1. DNA replication-coupled repair of inter-strand DNA crosslinks. (A) NEIL1 excise unhooked crosslinked oligomer in three-stranded DNA repair/replication intermediate. (B) NEIL3- and (C) Fanconi anemia-mediated repair of ICL. (D) The hypothetical mechanism of the FANCM-mediated DNA replication fork bypass of ICL and NEILs-mediated repair of three- and four stranded crosslinked DNA structures.
Axis 2: Study of new post-replicative modifications of DNA by poly(ADP-ribose) polymerases (PARPs) and their role in the repair of DNA strand breaks in normal and cancer cells.
PARP enzymes are major players in DNA damage response and repair of DNA strand breaks. Over-activation of PARP1 is often associated with drug resistance of cancer cells. Noteworthy, PARP1 interacts with HIF and regulates the fine tuning of the HIF-mediated hypoxic response in vivo. In our previous studies we demonstrated that:
- The cisplatin-resistant cancer cells manifest hyper-activation of PARP1 and increased cell death when exposed to PARP1-inhibitors, as compared to cisplatin-sensitive cancer cells (Michels et al., 2013);
- Human PARP1, 2 and 3 proteins can covalently poly- and mono-ADP-ribosylate DNA duplex extremities under in vitro conditions (Belousova et al, 2018; Talhaoui et al, 2016; Zarkovic et al, 2018) (see Figure 2);
- PARPs catalysed covalent ADP-ribosylation of DNA is fully reversible process since this modification is rapidly degraded by Poly(ADP-ribose) glycohydrolase (PARG) (Talhaoui et al, 2016).
Our preliminary data suggest that the covalent ADP-ribosylation of DNA termini occur as a cellular response to complex DNA damage and may be involved in the coordination of repair of multiple closely spaced DNA strand breaks. The current research activities aim:
- to characterize mechanisms of DNA ADP-ribosylation by PARPs;
- to identify proteins responsible for sensing of ADP-ribosylated DNA adducts;
- to search for the presence of ADP-ribosylated DNA adducts in cancer cells exposed to various genotoxic treatments;
- to identify the role of DNA ADP-ribosylation in the DNA damage responses and coordination of DNA strand break repair in normal and cancer cells, as well as in the acquired resistance of latter to chemotherapy.
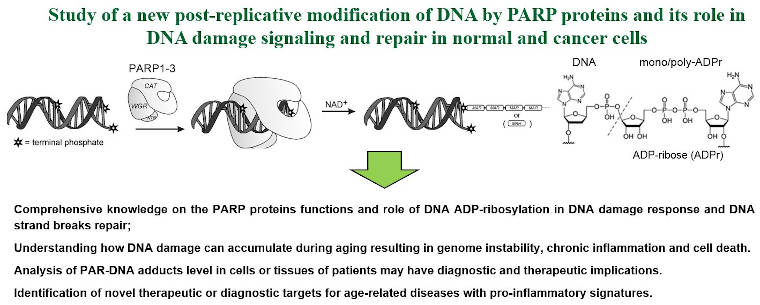
Figure 2. Axis 2: Study of a new post-replicative modification of DNA by PARP proteins.
Axis 3: Study of the signaling pathways induced by DNA damage in renal cancer and their associations with hereditary cancer syndromes.
Although most cases of renal cell carcinoma (RCC) seem to occur sporadically, an inherited predisposition accounts for about 3% of cases. Extensive studies of families affected with inherited renal cancers led to the identification of 4 main predisposing genes: 3 tumor suppressor genes VHL, FLCN, and FH and 1 oncogene, MET. In addition, we and others identified germline mutations in new genes: MITF, BAP1 and PBRM1 (Bertolotto et al, 2011; Popova et al, 2013; Benusiglio et al, 2015) (see Figure 3).
Identification of the causal mutation of RCC predisposing genes in kindreds offers the possibility of genetic testing and allows surveillance of mutation carriers resulting in earlier diagnosis and better prognosis. Furthermore, most of the predisposing genes are mutated in somatic cells in sporadic RCC, providing insights into the various mechanisms of renal tumorigenesis. This is especially the case with the VHL gene, also inactivated in most of the sporadic ccRCC. The pVHL is pivotal in angiogenesis: it targets the HIF for degradation under normal oxygen conditions. The discovery of VHL and its role helped to develop new drugs to inhibit HIF and its downstream targets, such as the anti-angiogenic treatments. Renal cancer is now considered to be a metabolic disease as major genes are involved in pathways converging towards the activation of HIF.
The French National Network for Rare Cancers in Adults “PREDIR”, chaired by Prof. S. Richard, is in charge of the genetic diagnostics and clinical managements of the patients with proven or suspected inherited kidney cancers. It allows us to access after informed consent to tumors samples and family blood samples. We revealed a gradient of pVHL dysfunction in hypoxia signalling pathways (Couvé et al, 2014) and deregulation of the miRNA and mRNA profiles in VHL-associated renal tumours (Gattolliat et al, 2018).
Recently, we have identified two missense germline mutations in two different families affected with hereditary RCC in a gene coding a DNA repair protein involved in the removal of ICLs, telomere maintenance and Fanconi anemia pathway. Our current project aims to characterize mutated forms of this gene at the cellular and tissue levels (in normal and kidney tumour samples) and to evaluate the functional consequences of these mutations after exposure of the cells to genotoxic drugs. Using samples from patients with hereditary CCR (blood and tumour biopsies), we are looking for novel mutations in genes involved in the response to DNA damage to obtain insights into molecular mechanisms of DNA repair associated with carcinogenesis and chemo-/radio-resistance of tumour cells, particularly of kidney cancer.

Figure 3. Renal Cell Carcinoma (RCC): mutated genes, intratumoral hypoxia and chemoresistance. The VHL gene is the main predisposing gene to hereditary RCC. Its germline mutations predispose to von Hippel-Lindau disease. This gene is also found mutated at somatic level in the majority of sporadic RCC. Recently, other genes were found mutated such as PBRM1, BAP1 and SETD2. PBRM1 is a member of the SWI-SNF chromatin remodelling complex and recent articles established a direct link between PBRM1 and HIF. BAP1 is a deubiquitinase for which a role was reported in repairing damaged DNA. SETD2 is a histone methyltransferase that has been shown recently to regulate DNA Mismatch Repair (MMR). Interestingly, they are all located in chromosome 3p, which is generally deleted in all renal tumors with a loss of heterozygosity (LOH).
